Welcome to ADAP-KDB Spectral Knowledgebase!
Use this web app to search query spectra against both private and public compounds and mass spectra.
Use it to prioritize both known and unknown mass spectra.
Use it as a search engine to identify spectra from libraries.
If you find ADAP-KDB useful, please cite
Smirnov A, Liao Y, Fahy E, Subramaniam S, Du X*: ADAP-KDB: A Spectral Knowledgebase for Tracking and Prioritizing Unknown GC-MS Spectra in the NIH's Metabolomics Data Repository.
Anal Chem 2021, 93(36):12213-12220
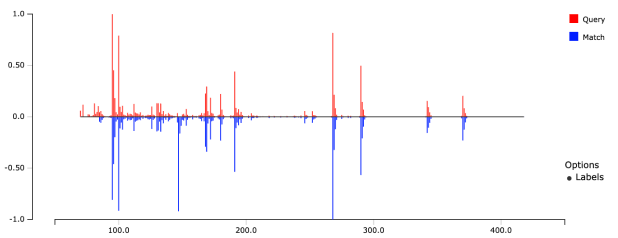
- Upload files
- Search for similar spectra and compounds
- Export results
- Upload files
- Save as a private library (user account required)
- Search against your libraries

- Search against ADAP-KDB consensus spectra
- Prioritize matches based on species/source/treatment distributions
- Find linked studies from Metabolomics Workbench
ADAP Spectral Knowledgebase is currently being actively developed by the Du-Lab team ( https://www.du-lab.org/).
If you encounter any issues or would like to provide a feedback on your experience using ADAP Spectral Knowledgebase click here, or please contact us through the email dulab.binf@gmail.com.